Post-processing LakeEnsemblR
Source:vignettes/articles/post-processing-lakeensemblr.Rmd
post-processing-lakeensemblr.RmdOnce LakeEnsemblR did successfully run all lake models, you can either extract the output data from the netcdf file for custom analysis and plotting, or use post-processing scripts for an initial look at the output.
Quick post-processing
# define path to the LakeEnsemblR netcdf output
ens_out <- "output/ensemble_output.nc"
# Plot depth and time-specific results
p <- plot_ensemble(ncdf = ens_out, model = c('FLake', 'GLM', 'GOTM', 'Simstrat', 'MyLake'), depth = 0.9,
var = 'watertemp', date = as.POSIXct("2010-06-13", tz = "UTC"), boxwhisker = TRUE, residuals = TRUE)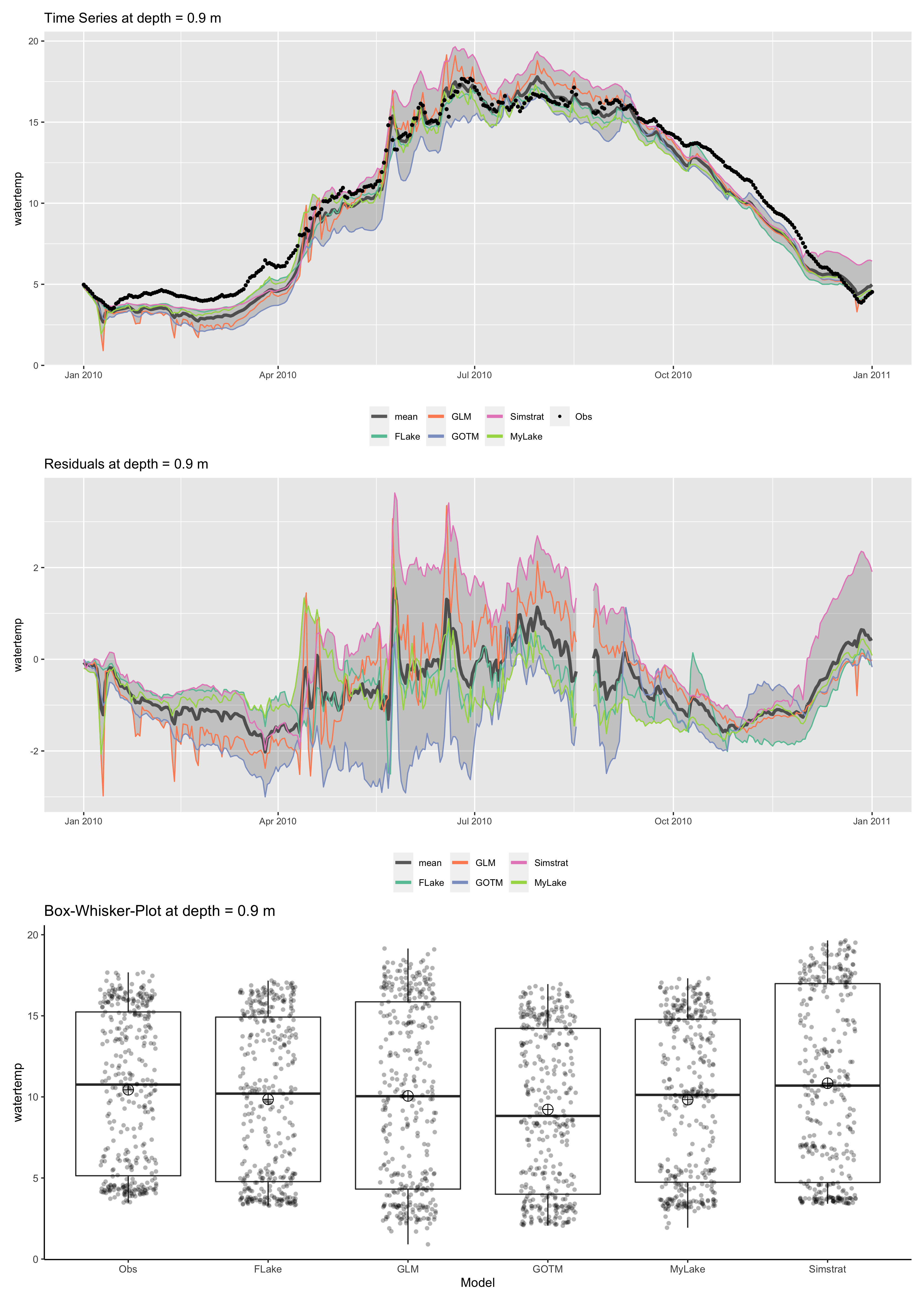

# Take a look at the model fits to the observed data
calc_fit(ncdf = "output/ensemble_output.nc",
model = c("FLake", "GLM", "GOTM", "Simstrat", "MyLake"),
var = "watertemp")
print(calc_fit)
$FLake
rmse nse r re nmae
1 3.356338 0.651542 0.6452533 -0.1836044 0.1877008
$GLM
rmse nse r re nmae
1 2.677758 0.5880857 0.8791993 -0.2142704 0.2230113
$GOTM
rmse nse r re nmae
1 2.855198 0.5303783 0.8983338 -0.2464723 0.2478081
$Simstrat
rmse nse r re nmae
1 4.111067 0.02639042 0.653375 -0.2212801 0.2915025
$MyLake
rmse nse r re nmae
1 2.548408 0.6258777 0.9059941 -0.1880694 0.1946865
# Take a look at the model performance against residuals, time and depth
plist <- plot_resid(ncdf = ens_out,var = "watertemp",
model = c('FLake', 'GLM', 'GOTM', 'Simstrat', 'MyLake'))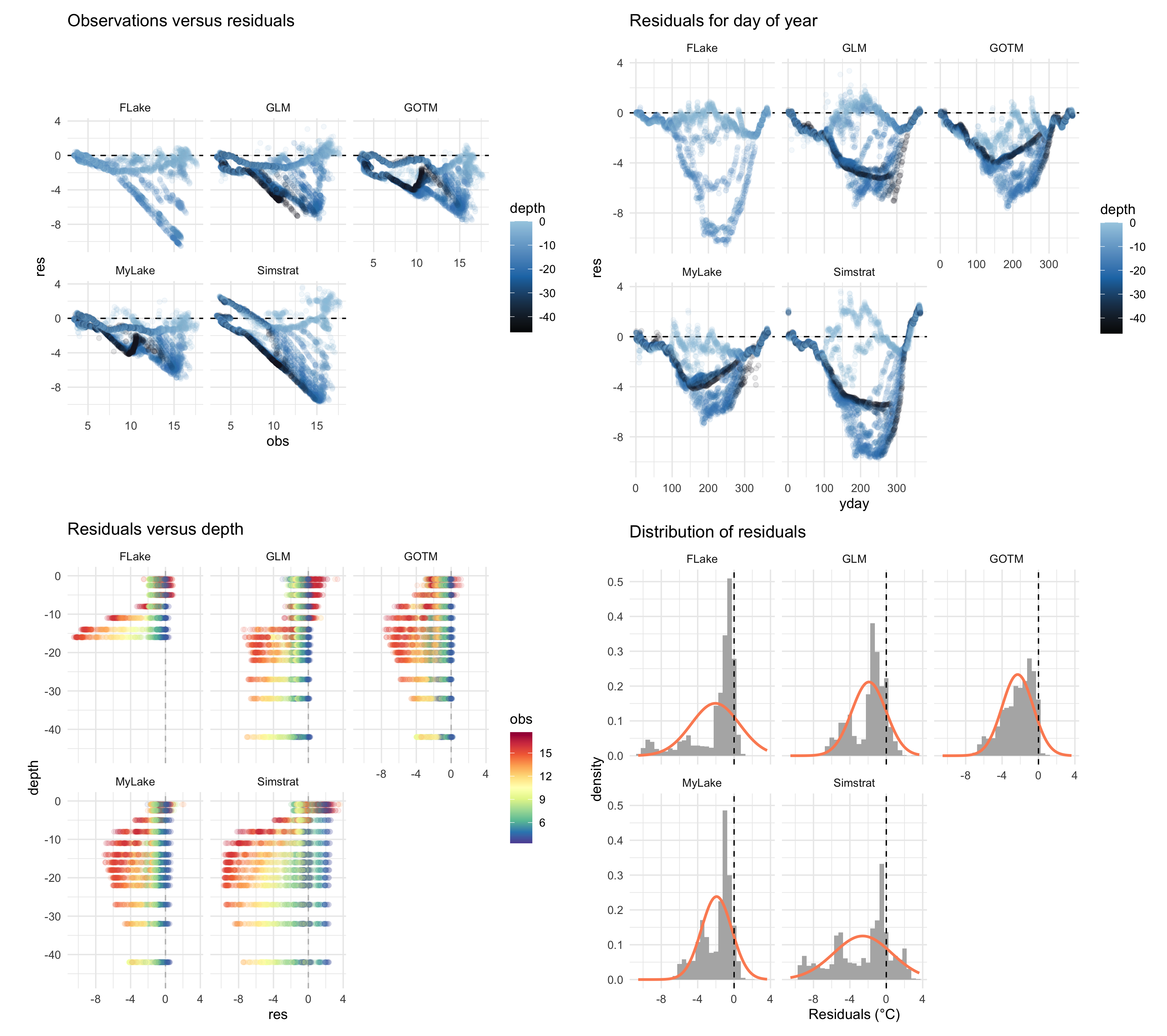
Further custom post-processing
# Load post-processed output data into your workspace
analyse_df <- analyse_ncdf(ncdf = ens_out, spin_up = NULL, drho = 0.1)
# Example plot the summer stratification period
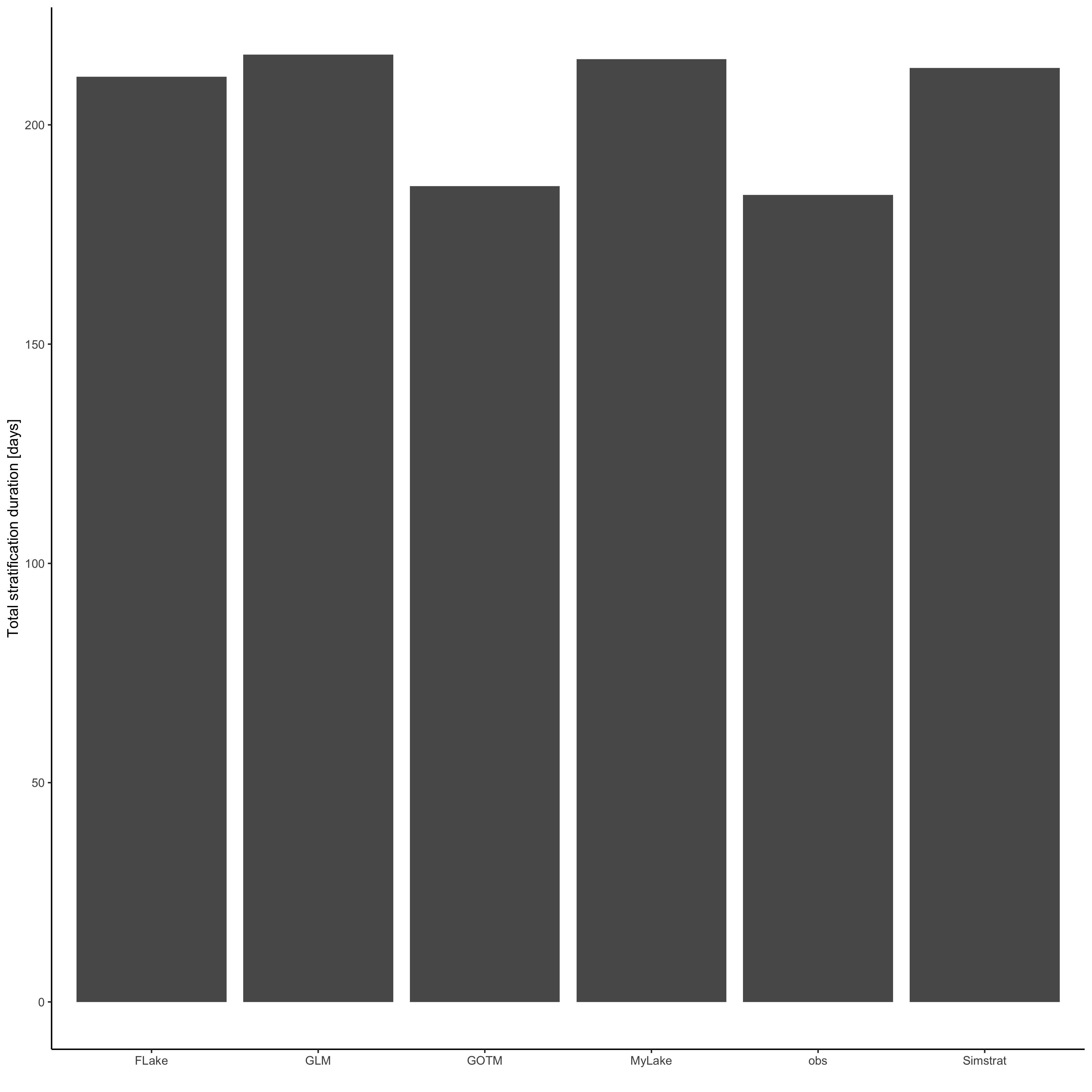
strat_df <- analyse_df$strat
p <- ggplot(strat_df, aes(model, TotStratDur)) +
geom_col() +
ylab("Total stratification duration [days]") +
xlab("") +
theme_classic()
ggsave("output/model_ensemble_stratification.png", p, dpi = 300, width = 284, height = 284, units = "mm")